Building an Extensible, rEproducible, Test-driven, Harmonized, Open-source, Versioned, ENsemble model for air quality 
Group Project for the Spatiotemporal Exposures and Toxicology group with help from friends 😃 🤠 🌎
Installation
remotes::install_github("NIEHS/beethoven")Workflow
beethoven is a targets reproducible analysis pipeline with the following workflow.
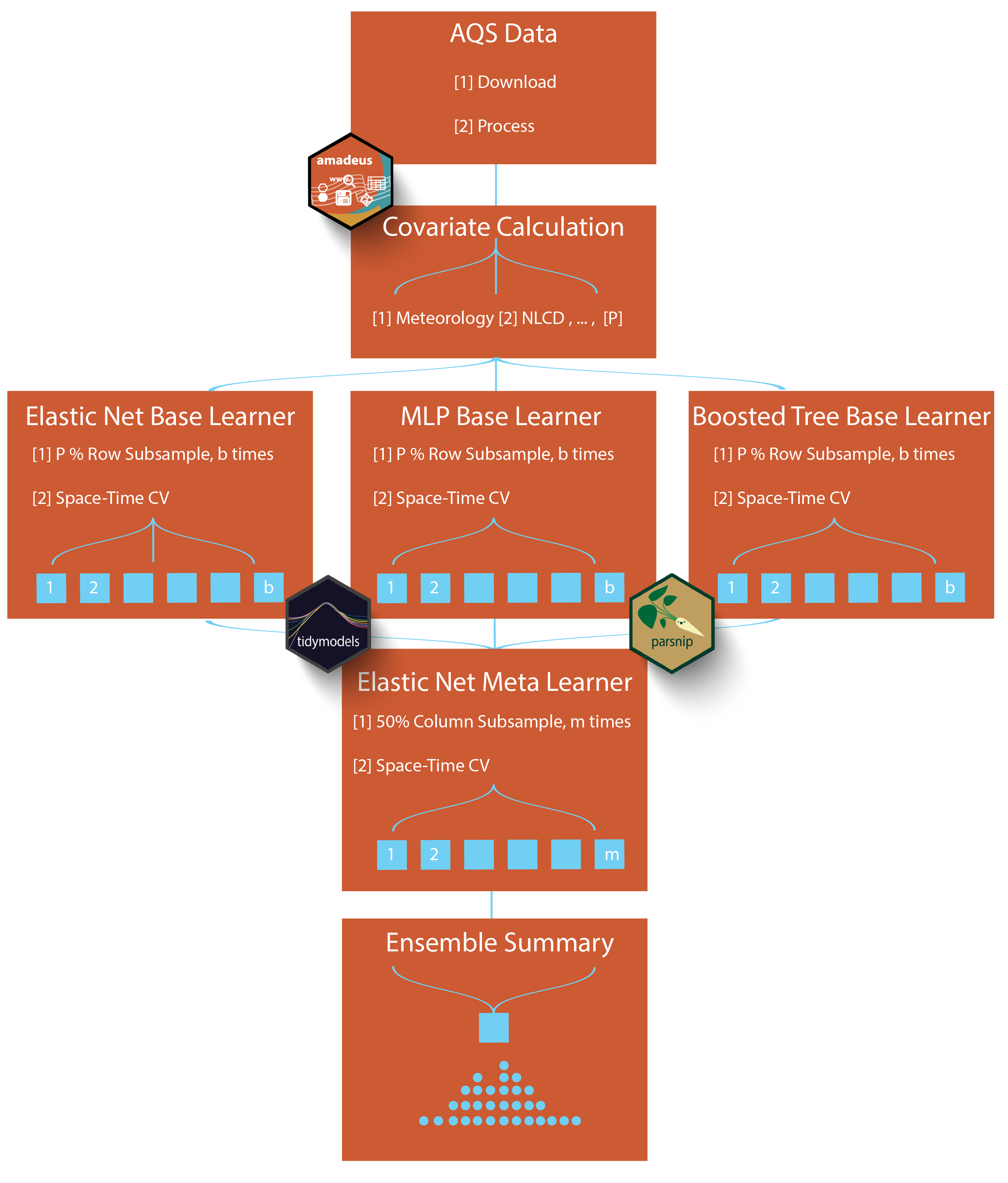
beethoven workflowVersion 0.4.4 of beethoven has stable targets for downloading data files, calculating features at AQS sites, and merging to a base learner-ready data.table (dt_feat_calc_xyt). Ongoing changes relate to calculating features for the prediction grid, computationally managing prediction grid, base learner hyperparameter tuning, and meta learner function development.
targets::tar_visnetwork()
beethoven targets::tar_visnetwork()Organization
Here, we describe the structure of the repository, important files, and the targets object naming conventions.
Folder Structure
-
R/is where thebeethovenfunctions are stored. Only “.R” files should be in this folder (ie.targetshelpers, post-processing, model fitting functions). -
inst/is a directory for arbitrary files outside of the mainR/directory-
targets/is a sub-directory withininst/which contains the pipeline files (ie. “targets_aqs.R”). These files declare thetargets::tar_targetobjects which constitute thebeethovenpipeline.
-
-
tests/stores unit and integration tests (testthat/) and test data (testdata/) according to the testthat package’s standard structure. for unit testing.-
testthat.Ris created and maintained bytestthat, and is not to be edited manually.
-
-
container/stores definition files and build scripts to build covariate- and model-specific Apptainer container images (container_covariates.defandcontainer_models.def). -
man/contains function documentation files (“.Rd”) which are by the roxygen2 package. These files are not to be edited manually. -
vignettes/contains “.Rmd” narrative text and code files. These are rendered by pkgdown into the Articles section of thebeethovenwebpage. -
.github/workflows/is a hidden directory which stores the GitHub CI/CD “yaml” files. -
tools/is dedicated to educational or demonstration material (e.g. Rshiny), but is not excluded from the package build.
Important Files
-
_targets.Rconfigurestargetssettings, creates computational resource controllers, and structures thebeethovenpipeline.- To run
beethoven, users must review and update the following parameters for their user profile and computing system:-
controller_*Ensure the local controllers do not request more CPUs than are available on your machine or high performance system. -
#SBATCH --partitionUtilization of NVIDIA GPUs (withinglue::gluecommand) -
--bind /USER_PATH_TO_INPUT/input:/input(withinglue::gluecommand)
-
- To run
-
_targets.yamlis created and updated by runningtargets::tar_makeand is not to be edited manually. -
run.shsubmits separateSBATCHjobs for the covariate, cpu- and gpu-enabled base learner, and the meta learnertargets(see/inst/scripts/). This setup ensures that each stage utilizes the proper container image and computational resources. To runbeethoven, users must review and update the following parameters for their user profile and computing system in each of theinst/scripts/run_*files.:#SBATCH --mail-user#SBATCH --partition#SBATCH --mem#SBATCH --cpus-per-task--bind /USER_PATH_TO_INPUT/input:/input--bind /USER_PATH_TO_SLURM/slurm:/USER_PATH_TO_SLURM/slurm
Running beethoven Pipeline
User settings
beethoven pipeline is configured for SLURM with defaults for NIEHS HPC settings. For adapting the settings to users’ environment, consult with the documentation of your platform and edit the requested resources in the stage-specific run files (/inst/scripts/) (lines 3-11) and _targets.R (lines 41-45; individual crew and crew.cluster controller workers).
Critical targets
There are 5 “critical” targets that users may want to change to run beethoven.
-
chr_daterange- Controls all time-related targets for the entire pipeline. This is the only
targetthat needs to be changed to update the pipeline with a new temopral range. Month and year specific arguments are derived from the time range defined bychr_daterange.
- Controls all time-related targets for the entire pipeline. This is the only
-
chr_nasa_token- Sets the file path to the user’s NASA Earthdata account credentials. These credentials expire at ~90 day intervals and therefore must be updated regularly.
-
chr_mod06_links- The file path to the MOD06 links file. These links must be manually downloaded per the
amadeus::download_modisfunction. The links are then stored in a CSV file that is read by the function. The new file with links must be updated to match the new date range.
- The file path to the MOD06 links file. These links must be manually downloaded per the
-
chr_input_dir- The file path to the input directory. This target controls where the raw data files are downloaded to and imported from. This file path must be mounted to the container at run time in the
run.shscript.
- The file path to the input directory. This target controls where the raw data files are downloaded to and imported from. This file path must be mounted to the container at run time in the
-
num_dates_split- Controls the size of temporal splits. Splitting the temporal range into smaller chunks allows for parallel processing across multiple workers. It also allows for dispatching new dynamic branches when the temporal range is updated.
Apptainer
Current implementation of beethoven utilizes Apptainer images to run the pipeline with consistent package versions and custom installations. Users must build these images before runnning beethoven.
cd container/ # must be working in the `container/` directory
sh build_container_covariates.sh # build "covariates" stage image
sh build_container_models.sh # build "models" image
mv *sif ../ # move images to `beethoven/` root directory[!NOTE]
.siffiles are omitted from GitHub due to size (>5 Gb each)
Run
After switching back to the project root directory, users can run the pipeline with the run.sh shell script. The following lines of /inst/scripts/run_*.sh must be updated with user-specific settings before running the pipeline
#SBATCH --mail-user=[USER_EMAIL] # email address for job notifications
#SBATCH --partition=[PARTITION_NAME] # HPC partition to run on
#SBATCH --mem=[###G] # Total memory for the job
#SBATCH --cpus-per-task=[###] # Total CPUs for the job
...
--bind [USER_INPUT_DIRECTORY]/input:/input \
...
--bind [USER_SYSTEM_PATH/munge]:/run/munge \
--bind [USER_SYSTEM_PATH/slurm]:[USER_SYSTEM_PATH/slurm] \Once configured, the pipeline can be run with a SLRUM batch job.
The SLURM batch job can also be submitted R session with the batch helper function.
source("R/helpers.R")
batch()Contribution
The Developer’s Guide provides detailed instructions for how to develop or update beethoven settings or individual targets objecdts
To contribute developments or modifications, open a Pull request into the dev branch with a detailed description of the proposed changes. Pull requests must pass all status checks, and then will be approved or rejected by beethoven’s authors.
Utilize Issues to notify the authors of bugs, questions, or recommendations. Identify each issue with the appropriate label to help ensure a timely response.
